
Microorganisms play a critical role in the food industry. Desirable yeasts and bacteria contribute to the production of foods and food ingredients. Undesirable yeasts and bacteria lead to food spoilage or worse, food safety risks.
Our aim is to understand how microorganisms interact with the medium, how they respond to environmental stress, and how they respond to the presence of other species. To do so, we use a multiscale modeling perspective: from biochemical networks inside the cell to microbial communities.
Models provide fundamental knowledge on how biological systems behave, but they can also be used to optimize biological systems and bioprocesses of interest in the food and aquaculture industries. Therefore, we put emphasis on developing methods and tools to facilitate modeling and systematizing processes and systems design, optimization, and control.
Modeling microbial systems at multiple scales
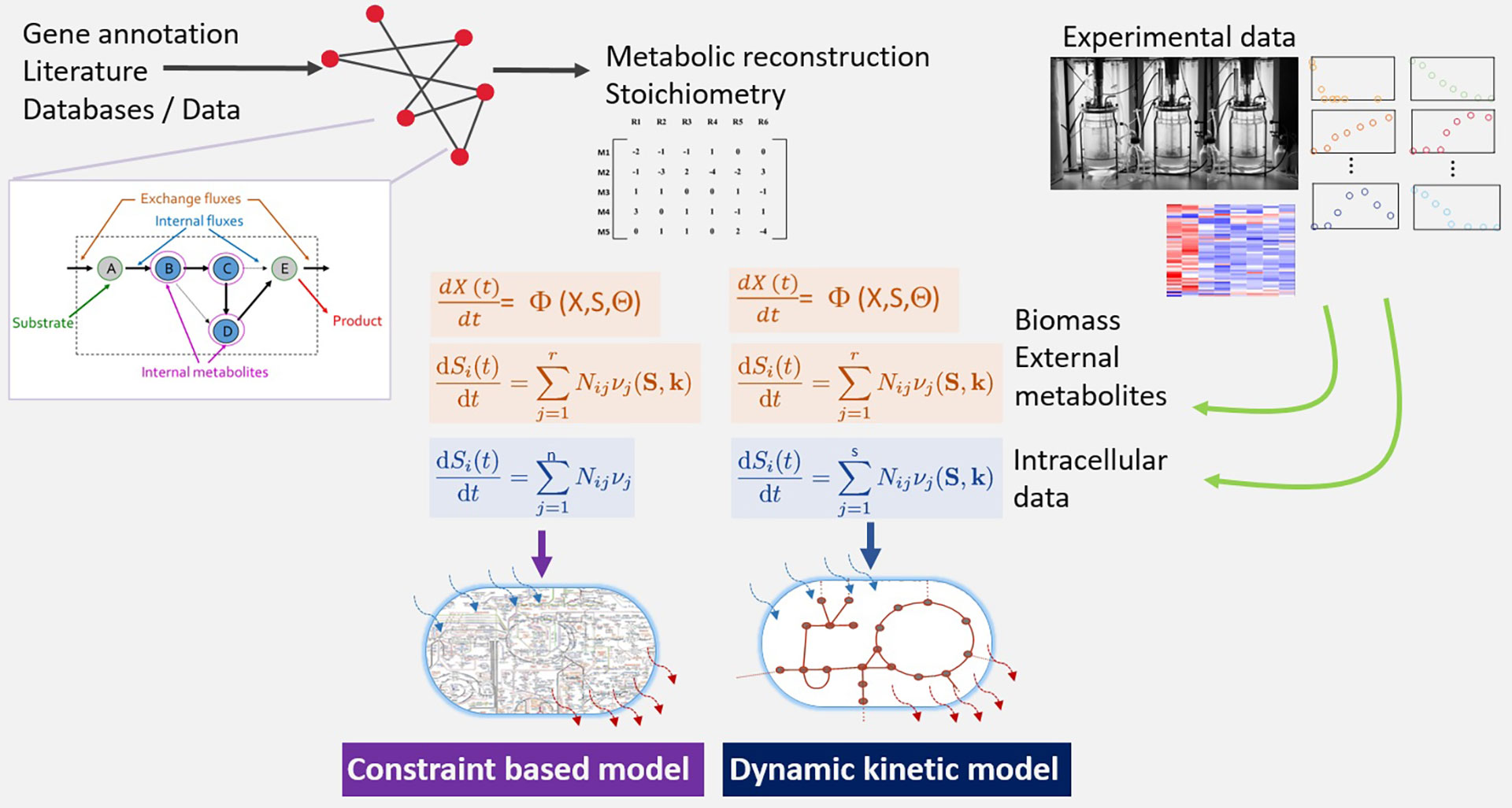
Currently, we focus on understanding the dynamics of growth and metabolism of microorganisms in fermentation conditions. To do so we use two types of models: kinetic coarse-grained models and detailed genome-scale models.
Genome-scale models are structured representations of a target organism based on existing genetic, biochemical, and physiological information. Dynamic versions provide information on the fluxes through metabolic pathways.
Kinetic models account for the dynamics of metabolite concentration and regulatory effects. Kinetic (also called dynamic or mechanistic) models of metabolic pathways are based on a set of nonlinear deterministic ordinary differential equations which quantitatively describe the system and predict the response to different inputs.
Recent applications include:
– Modeling of yeast fermentation in reach media
Modeling intereactions in biological systems
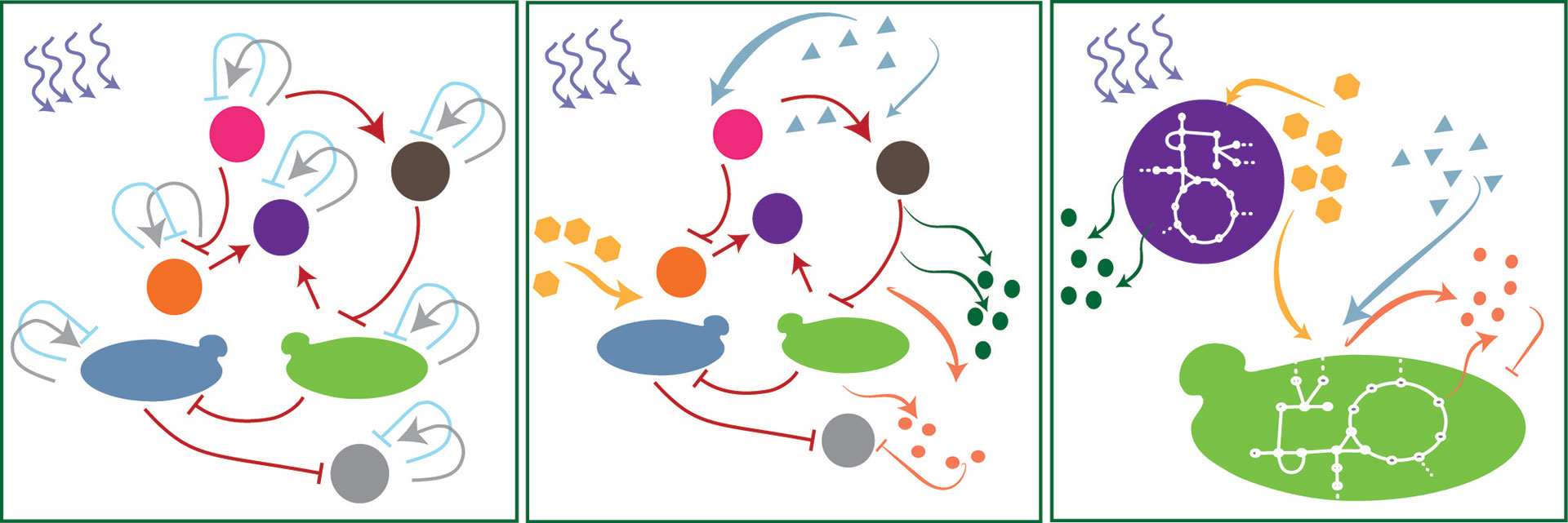
We use modeling to decipher the underlying ecological interactions between species. Interactions are governed by processes that occur at multiple scales, from molecules and cells to interacting populations and changing environments. To better understand and predict these processes, we need mechanistic models that can integrate information from simpler levels and generate accurate predictions at the community and system levels. As a first approximation, we consider classical ecological models; these models require information on the dynamics of the abundance of the species. We use more detailed models, yet coarse-grained, to understand the role of specific mechanisms such as competition for nutrients, cross-feeding, or exclusion due to the production of toxic compounds. Finally, we will approach more detailed, multi-omics models to better understand how cells use different metabolic pathways in the presence of other species.
Recent applications include:
– Modeling of Listeria monocytogenes biofilms formation
– Modeling multi-species in cheese fermentation
– Modeling of algae-probiotics interactions for aquaculture applications
Mathematical methods and software tools
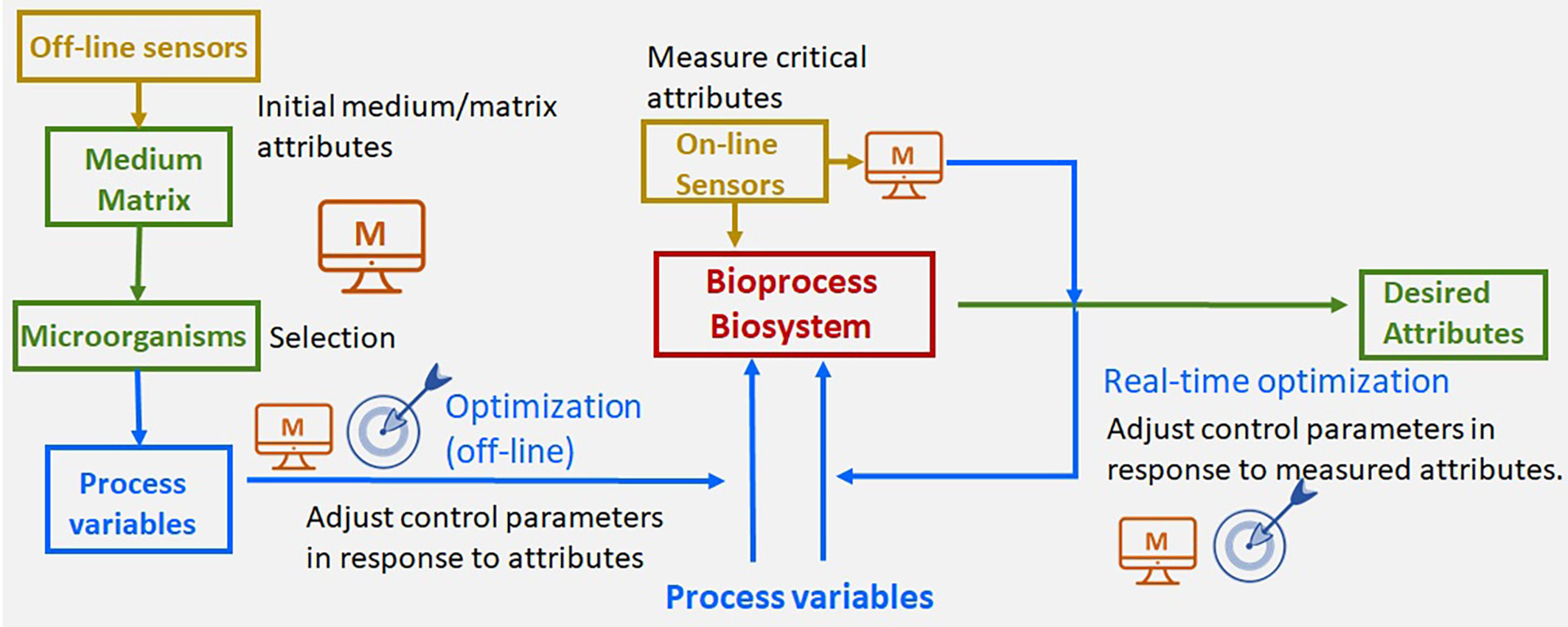
The success of mathematical models relies on their predictive capabilities. However, models often depend on, typically, unknown parameters. Therefore, parameter estimation, identifiability analysis, and optimal experimental design are critical to achieving the desired predictive properties. Predictive models can also be used to optimize or control biological systems and bioprocesses. Over the years, we have developed methods and tools to systematize modeling and optimization.
AMIGO2 solves (multi-objective) optimization problems at the core of systems biology: in model parametric identification; as the underlying hypothesis for model development and in the optimal control of biological systems to achieve the desired behavior.
GenSSI2 enables the multi-experiment structural identifiability analysis of non-linear dynamic models.
Applications in monitoring, design, optimization and control
Funding


FAIROMICS: Fairification of multiomics data to link databases and create knowledge graphs.

e-MUSE: Complex microbial Ecosystems MUltiScale modElling: mechanistic and data driven approaches integration

DTWINE: Digital twins to optimize energy efficiency and product quality in wineries
DIGITALG: Programa Ciencias Marinas, Gemelo digital del cultivo de algas en acuicultura multitrófica integrada
SENSWINE: Sensores inteligentes y predictivos para la monitorización rápida de la calidad de vino.

Ayuda de Grupos de Investigación
Contrato programa

